
Determination of the protein content of complex samples by aromatic amino acid analysis, liquid chromatography-UV absorbance, and colorimetry | SpringerLink
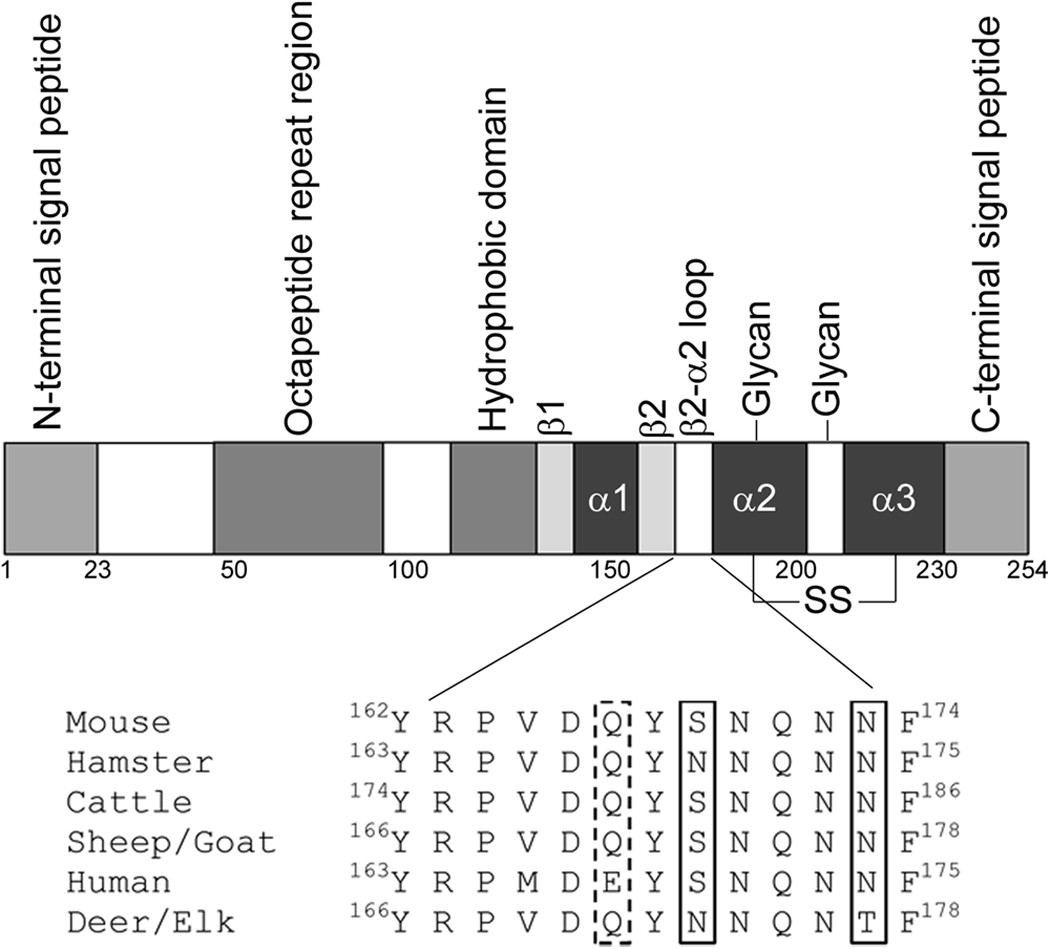
Complex folding and misfolding effects of deer-specific amino acid substitutions in the β2-α2 loop of murine prion protein | Scientific Reports

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal

Photometric Quantification of Proteins in Aqueous Solutions via UV-Vis Spectroscopy - Eppendorf Handling Solutions
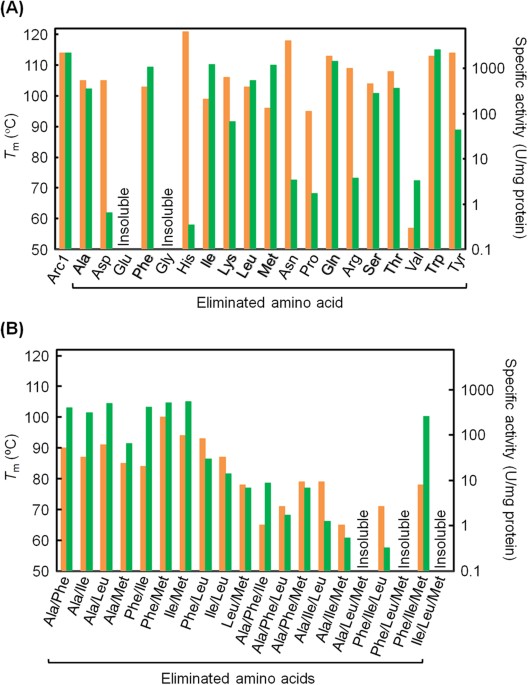
Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins | Scientific Reports
A point mutation in the microtubule binding region of the Ncd motor protein reduces motor velocity. - Abstract - Europe PMC

Artificial intelligence and machine learning for protein toxicity prediction using proteomics data - Vishnoi - 2020 - Chemical Biology & Drug Design - Wiley Online Library
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/2-Table1-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

SOLVED: It is possible to estimate the molar extinction coefficient of a protein from knowledge of its amino acid composition, as shown from your experiences with ExPASY: From the molar extinction coefficient
Comprehensive mutagenesis to identify amino acid residues contributing to the difference in thermostability between two originally thermostable ancestral proteins | PLOS ONE

Molar absorption coefficients for Trp, Tyr, and cystine based on an... | Download Scientific Diagram

How to measure and predict the molar absorption coefficient of a protein - Pace - 1995 - Protein Science - Wiley Online Library

Observed and predicted molar absorption coefficients at 280 nm for 80... | Download Scientific Diagram

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal
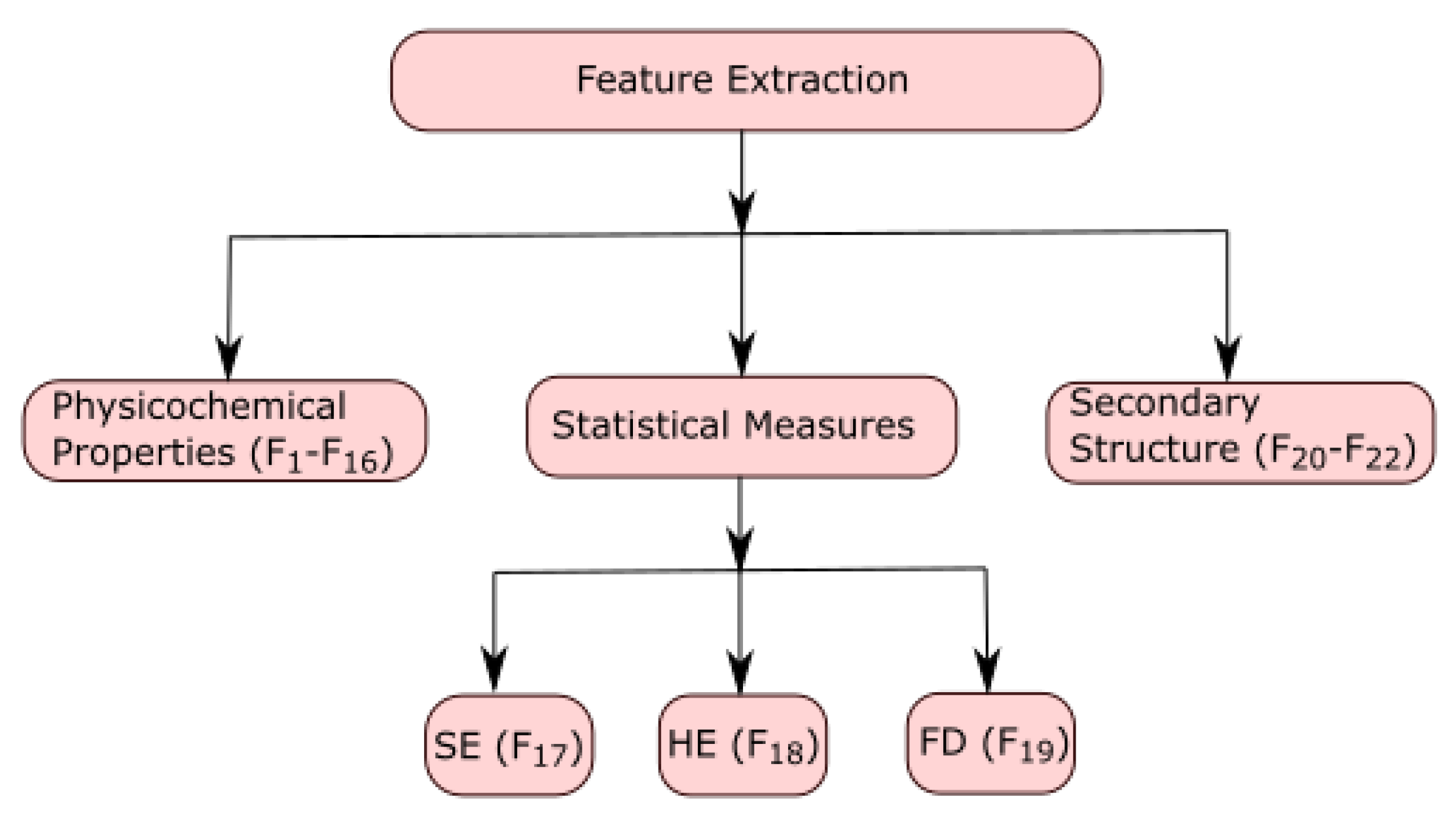
Mathematics | Free Full-Text | Unsupervised Learning for Feature Representation Using Spatial Distribution of Amino Acids in Aldehyde Dehydrogenase (ALDH2) Protein Sequences







